
Get Amino Acid Position from PDB Files: A Detailed Guide for You
Understanding the structure of proteins is crucial in biochemistry and molecular biology. One of the most common ways to study protein structures is by analyzing Protein Data Bank (PDB) files. These files contain detailed information about the three-dimensional arrangement of atoms in a protein. In this article, I will guide you through the process of extracting amino acid positions from PDB files using Python. By the end of this article, you will be able to analyze PDB files like a pro!
Understanding PDB Files
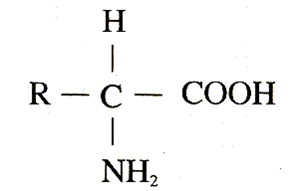
PDB files are text files that contain atomic coordinates, bond information, and other relevant data about a protein. Each PDB file is identified by a unique four-character code. The file format is based on the mmCIF (macromolecular Crystallographic Information File) standard, which is a data exchange format for macromolecular crystallographic data.
When you open a PDB file, you will see a series of records, each starting with a line that begins with a specific character. For example, the “ATOM” record contains the coordinates of an atom, while the “HETATM” record contains the coordinates of a non-standard atom. The “AUTHOR” record provides information about the authors of the PDB entry, and the “REMARK” record contains comments or other information about the protein.
Setting Up Your Python Environment

Before you start extracting amino acid positions from PDB files, you need to set up your Python environment. Make sure you have Python installed on your computer. You will also need to install the following packages:
You can install these packages using pip:
pip install requests requests-cache pandas MDAnalysisExtracting Amino Acid Positions
Now that you have your Python environment set up, let’s dive into the code. We will use the MDAnalysis library to read the PDB file and extract the amino acid positions.
Here’s a sample code snippet to get you started:
from MDAnalysis import Universe Load the PDB filepdb_file = "1A3N.pdb"u = Universe(pdb_file) Get the atom selection for amino acidsselection = u.select_atoms("resname ALA") Print the amino acid positionsfor atom in selection.atoms: print(f"Atom name: {atom.name}, Position: {atom.position}")In this code, we first import the Universe class from the MDAnalysis library. Then, we load the PDB file using the Universe constructor. Next, we create a selection for amino acids with the “resname ALA” expression, which selects all atoms with the residue name “ALA”. Finally, we iterate over the selected atoms and print their names and positions.
Handling Multiple Amino Acids
When analyzing a protein, you may want to extract positions for multiple amino acids. You can modify the selection expression to include multiple residue names, like this:
selection = u.select_atoms("resname ALA, GLY, SER")This will select all atoms with the residue names “ALA”, “GLY”, or “SER”. You can add as many residue names as you need to the selection expression.
Visualizing Amino Acid Positions
Visualizing the amino acid positions can help you better understand the protein structure. You can use the MDAnalysis library to plot the positions of the selected atoms.
Here’s an example code snippet to plot the amino acid positions:
import matplotlib.pyplot as plt Get the amino acid positionspositions = [atom.position for atom in selection.atoms] Plot the positionsplt.scatter(positions[:, 0], positions[:, 1], c='blue', marker='o')plt.xlabel("X-axis")plt.ylabel("Y-axis")plt.title("Amino Acid Positions")plt.show()In this code, we first extract the positions of the selected atoms and store them in a list. Then, we use the matplotlib library to create a scatter plot of the positions






