
Understanding BAM Files: A Comprehensive Guide
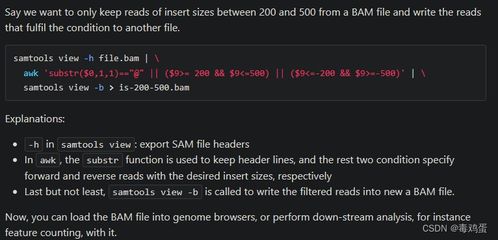
BAM files, or Binary Alignment/Map files, are a crucial component in the world of genomic data analysis. They are used to store large-scale sequencing data, making them an essential tool for researchers and scientists. In this article, we will delve into the intricacies of BAM files, exploring their structure, uses, and how they are processed. Let’s embark on this journey to uncover the mysteries of BAM files.
What is a BAM File?
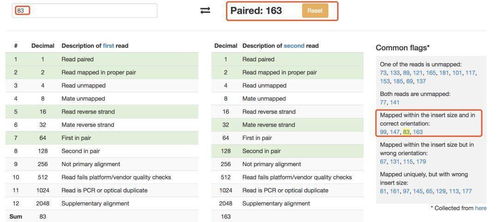
A BAM file is a compressed, indexed binary file format that represents mapped reads in a reference genome. It is designed to store large amounts of sequence data efficiently, making it an ideal choice for handling the vast quantities of data generated by next-generation sequencing technologies. BAM files are compatible with the SAM (Sequence Alignment/Map) format, which is a text-based format used for storing alignment data.
Structure of a BAM File
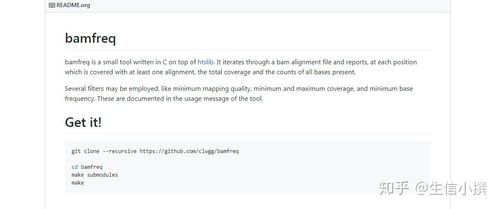
The structure of a BAM file is quite complex, but understanding it is essential for working with these files effectively. A BAM file consists of several components:
| Component | Description |
|---|---|
| Header | Contains metadata about the file, such as the reference genome, read groups, and alignment information. |
| Body | Stores the actual alignment data, including the read names, positions, and mapping quality scores. |
| Index | Enables quick access to specific regions of the genome by providing a map of the file’s contents. |
Understanding the structure of a BAM file is crucial for efficiently processing and analyzing the data it contains.
Uses of BAM Files
BAM files are widely used in various genomic analysis applications, including:
- Variant calling: Identifying genetic variations, such as single nucleotide polymorphisms (SNPs) and insertions/deletions (indels), in a sample.
- Genome assembly: Constructing a reference genome from a set of short reads.
- Expression analysis: Quantifying gene expression levels by counting the number of reads that map to a gene.
- Chromosome conformation capture (3C): Studying the spatial organization of chromosomes.
BAM files are an essential tool for researchers and scientists working in the field of genomics, as they enable the efficient processing and analysis of large-scale sequencing data.
Processing BAM Files
Processing BAM files often involves several steps, including alignment, sorting, indexing, and filtering. Here’s a brief overview of these steps:
- Alignment: Mapping reads to a reference genome using tools like BWA, Bowtie, or STAR.
- Sorting: Organizing the aligned reads in a specific order, such as by coordinate or read name, using tools like SAMtools or Picard.
- Indexing: Creating an index file that allows for quick access to specific regions of the genome, using tools like SAMtools or Picard.
- Filtering: Removing reads that do not meet certain criteria, such as mapping quality or read length, using tools like SAMtools or Picard.
These steps are essential for preparing BAM files for downstream analysis, such as variant calling or expression analysis.
Conclusion
BAM files are a powerful tool for genomic data analysis, enabling researchers and scientists to efficiently process and analyze large-scale sequencing data. Understanding the structure, uses, and processing steps of BAM files is crucial for working with these files effectively. By mastering the intricacies of BAM files, you’ll be well-equipped to tackle the challenges of genomic data analysis in your research.




